what components are necessary to synthesize dna in vitro?
1.iv: In vitro applications of DNA replication
- Folio ID
- 25725
Learning Objectives
- Compare cellular Deoxyribonucleic acid replication with the homo applications of PCR and sequencing methods.
- Explicate the part of primers, Taq polymerase, and the temperature cycles in PCR.
- Explain the human relationship betwixt the structure of dideoxynucleotides and their role in sequencing reactions.
- Depict the procedure of traditional (Sanger) sequencing.
- Give an example of a NextGen sequencing method and briefly explain how information technology differs from traditional sequencing.
Agreement and adapting cellular DNA replication has led to innovations that accept had an immeasurable bear on on research, biotechnology, and medicine. Both polymerase chain reaction (PCR) and Dna sequencing are in vitro applications of cellular Deoxyribonucleic acid replication.
PCR
Molecular cloning was the showtime method available to isolate a gene of involvement and make many copies of it to obtain sufficient amounts of the DNA to study. Today, there is a faster and easier manner to obtain large amounts of a Deoxyribonucleic acid sequence of interest: the polymerase chain reaction (PCR). PCR allows i to use the power of Dna replication to amplify DNA enormously in a short menstruum of time. As you know, cells replicate their Deoxyribonucleic acid before they divide, and in doing so, double the amount of the cell's Dna. PCR substantially mimics cellular DNA replication in the test tube, repeatedly copying the target Dna over and over, to produce large quantities of the desired Dna.
Selective Replication
In contrast to cellular Deoxyribonucleic acid replication, which amplifies all of a prison cell's Dna during a replication wheel, PCR does targeted amplification to replicate only a segment of DNA divisional by the two primers that make up one's mind where DNA polymerase begins replication. Each bicycle of PCR involves three steps, denaturing, annealing and extension, each of which occurs at a different temperature.

The Starting Materials
Since PCR is, basically, replication of Dna in a test-tube, many of the usual ingredients needed for Deoxyribonucleic acid replication are required:
- A template (the DNA containing the target sequence that is being copied)
- Primers (to initiate the synthesis of the new Deoxyribonucleic acid strands)
- Thermostable DNA polymerase (to acquit out the synthesis). The polymerase needs to be oestrus stable, because heat is used to separate the template DNA strands in each cycle.
- dNTPs (DNA nucleotides to build the new DNA strands).
- The template is the DNA that contains the target you desire to amplify (the "target" is the specific region of the DNA y'all want to amplify).
The primers are short constructed single-stranded DNA molecules whose sequence is complementary to a region flanking the target sequence. These Dna molecules can be chemically-synthesized to produce any given base sequence to use as primers. Primers are present in millions of fold excess over the template, because each new DNA strand will brainstorm from a primer. Therefore, information technology is necessary to know a fiddling bit of the template sequence on either side of the region of DNA to be amplified. DNA polymerases and dNTPs are commercially available from biotechnology supply companies.
- The first step of the process involves separating the strands of the target Dna by heating to near humid.
- Next, the solution is cooled to a temperature that favors complementary DNA sequences finding each other and making base pairs, a process called annealing. Since the primers are present in great excess, the complementary sequences they target are readily establish and base-paired to the primers. These primers direct the synthesis of Deoxyribonucleic acid. Just where a primer anneals to a DNA strand will replication occur, since DNA polymerases crave a primer to brainstorm synthesis of a new strand.
- In the third stride in the process, the Dna polymerase replicates Deoxyribonucleic acid past extension from the three' end of the primer, making a new Dna strand. At the end of the offset cycle, there are twice as many Deoxyribonucleic acid molecules, only every bit in cellular replication. But in PCR, steps 1-3 are repeated, usually for between 25 and xxx cycles. At the stop of the process, there is a theoretical yield of 230 (over 1 billion times) more DNA than there was to start. (This enormous amplification power is the reason that PCR is so useful for forensic investigations, where very tiny amounts of DNA may exist available at a crime scene.)
The temperature cycles are controlled in a thermocycler, which repeatedly raises and lowers temperatures according to the set up program. Since each cycle can be completed in a couple of minutes, the entire distension can be completed very rapidly. The resulting DNA is analyzed on a gel to ensure that it is of the expected size, and depending on what it is to be used for, may also be sequenced, to exist certain that information technology is the desired fragment.
Case \(\PageIndex{1}\)
PCR is used for ane type of exam for SARS-CoV-2, the virus associated with COVID-nineteen. Why would this tool exist helpful to notice the virus? What materials would be needed to perform this examination?
Solution
Because PCR exponentially amplifies the number of copies of genetic material, even when simply a pocket-sized amount of genetic material is nowadays, many copies tin be produced and detected. In this instance, a small amount of virus would be extremely hard to detect without PCR. However, this virus has an RNA (not DNA) genome, and then an extra footstep must convert RNA to DNA, and then copies can be made!
You would need: a sample of cells from an private to isolate genetic cloth from, Taq polymerase and advisable buffer, dNTPs, primers specific to the virus genetic sequence
DNA sequencing
Dna sequencing determines the social club of bases in a slice of Dna, a whole chromosome, or even a whole genome. Retrieve that Deoxyribonucleic acid obeys the rules of base pairing, and so determining the sequence of one strand ways that we know the other strand likewise. DNA sequencing is an awarding of DNA replication, because Dna strands are congenital using nucleotides or methods that allow the scientist to visualize which base of operations was incorporated at each position.
Dideoxy sequencing (also known as Sanger sequencing)
The original method adult by Sanger and others relied on variant nucleotides (dNTPs), dideoxynucleotides (ddNTPs), which are also known equally concatenation terminators. Remember, that Deoxyribonucleic acid synthesis ever proceeds from v' to 3' past attaching the phosphate of the incoming nucleotide to the 3' OH of the previous nucleotide. When that hydroxyl group is absent as in a dideoxynucleotide, synthesis ends.
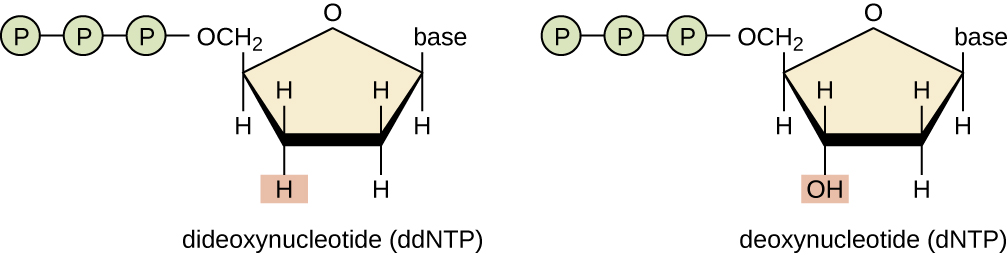
Figure \(\PageIndex{4}\): A dideoxynucleotide has a hydrogen on the three' carbon, whereas a deoxynucletide has a hydroxyl group. (https://cnx.org/contents/5CvTdmJL@4.four, CC BY iv.0, https://commons.wikimedia.org/w/inde...curid=53713264)
When a combination of dNTPs and ddNTPs are included, a mixture of DNA sequences of different lengths is produced. Each fragment ends with a labeled didexoynucledotide. Note that this process kickoff utilized radioactively-labeled nucleotides; merely one blazon of radioactively-labeled dideoxynucleotide (ddATP, ddGTP, ddTTP, or ddCTP) could be included in each reaction, meaning 4 individual reactions had to be performed. The utilise of fluorescent labels for the dideoxynucleotides simplified this process, allowing all four bases to be sequenced in a single reaction. Ordering the mixture of fragments by size using electrophoresis and reading the wavelength of each characterization using a light amplification by stimulated emission of radiation volition reveal the order of bases.
The reagents needed for a sequencing reaction include:
- Template Deoxyribonucleic acid (the DNA yous desire to know the sequence of)
- A mixture of all four deoxynucleotides (dATP, dCTP, dGTP, AND dTTP)
- Fluorescently-labeled dideoxynucleotide (ddATP, ddCTP, ddGTP, OR ddTTP)
- Deoxyribonucleic acid polymerase (and buffer for efficient enzyme role)
- Dna oligonucleotide primer (complementary to the sequence 5' of the unknown sequence)
Video \(\PageIndex{2}\): Run into how the mixture of fragments produced by this reaction can exist used to decide a sequence of DNA in this video. Note that whether a dNTP or ddNTP is incorporated at each position occurs at random. Past risk, all possible fragments lengths will be generated in a successful sequencing reaction. (Copyright SWLeacock own work)
Visualizing the sequence
After the sequencing reaction is completed, the randomly-terminated fragments are separated past size using electrophoresis. If the reaction proceeded optimally, the products are a mixture of fragments with each base represented at the end of a fragment. Each fragment ends with a fluorescently-labeled dideoxyribonucleotide. The smallest fragments will drift the fastest and pass by a laser that will detect the intensity of the fluorescence and record the corresponding base of operations. The results can be visualized in a plot that represents the peaks of fluorescence intensity at each size.
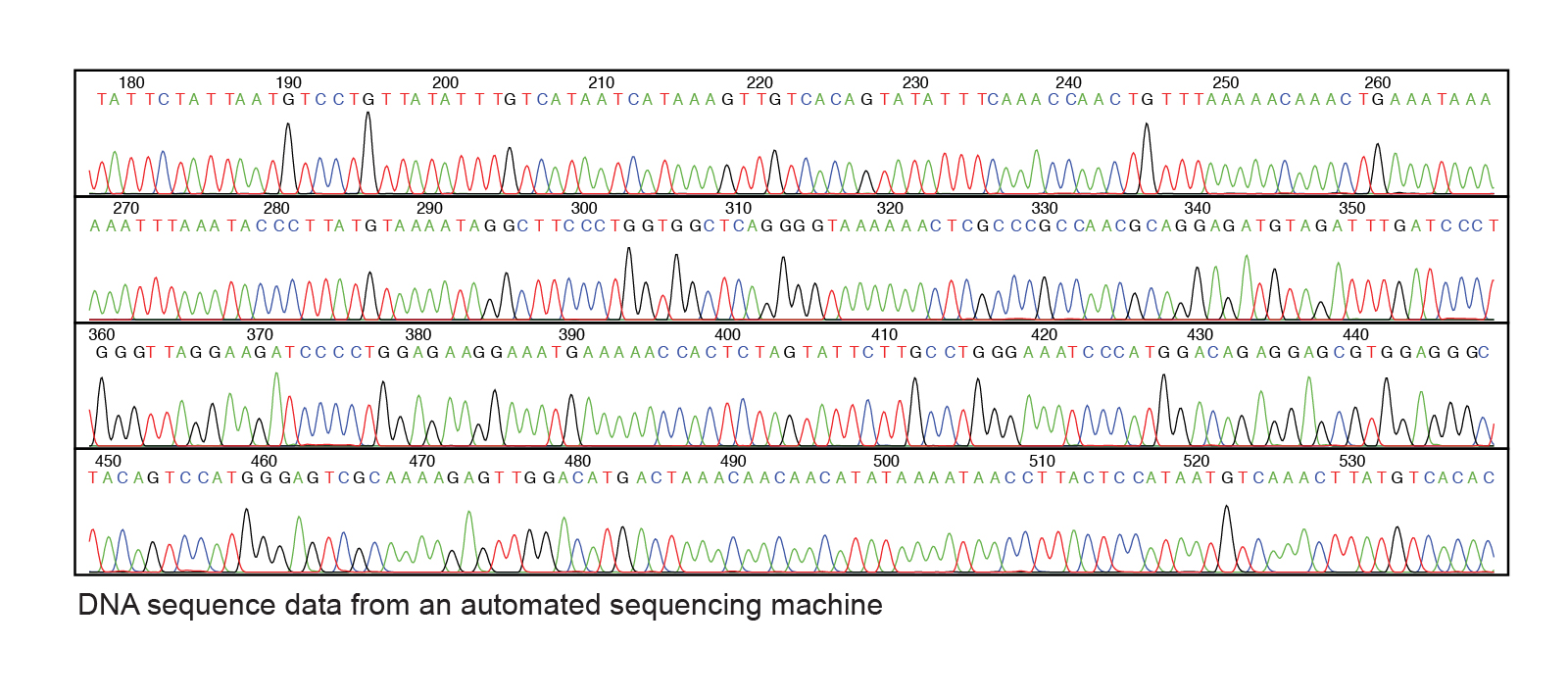
Practise \(\PageIndex{1}\)
one. Starting at the A at nucleotide 450 in the figure to a higher place, requite twenty nucleotides of sequence that recorded? (Bear witness the 5' and 3' ends.)
2. Based on your respond above, what was the sequence of the template DNA strand? (Show the 5' and 3' ends.)
- Answer
-
1. five' ACAGTCCATGGGAGTCGCAA iii'
two. 3' TGTCAGGTACCCTCAGCGTT five'
Next-generation sequencing
Traditional Sanger sequencing is however used for some sequencing applications, particularly for small pieces of Dna. However, newer technologies have improved the speed and lowered the cost of Deoxyribonucleic acid sequencing.
Three of these next-generation technologies (named for the companies that developed them) are highlighted hither, but others exist and new ones are likely.
Illumina Sequencing past Synthesis
Pacific Biosciences
Nanopore
Compare and contrast
- Compare and contrast cellular Deoxyribonucleic acid replication with DNA sequencing, addressing how Deoxyribonucleic acid sequencing depends on the rules of base pairing, Deoxyribonucleic acid structure, and Dna synthesis.
- Compare and contrast traditional sequencing and next-generation methods.
Do \(\PageIndex{1}\)
Which platform of next generation sequencing does not involve sequencing the DNA as it is existence synthesized by a Deoxyribonucleic acid polymerase?
A. Sanger sequencing
B. Illumina sequencing
C. Pacific Biosciences sequencing
D. Nanopore sequencing
- Respond
-
D. In Oxford Nanopore sequencing a double-stranded Deoxyribonucleic acid is separated and one strand is processed through the "pore" and changes in current are measured equally nucleotides pass through the aqueduct. Deoxyribonucleic acid polymerase is non synthesizing the strand while information technology is existence sequenced.
Source: https://bio.libretexts.org/Courses/University_of_Arkansas_Little_Rock/Genetics_BIOL3300_(Fall_2021)/Genetics_Textbook/01%3A_Chemistry_to_Chromosomes/1.04%3A_In_vitro_applications_of_DNA_replication